Parameters and Attributes
Input Parameters
Input parameter |
Description |
|---|---|
data |
(numpy.ndarray) n_samples x n_features. When using via_wrapper(), data is ANNdata object that has a PCA object adata.obsm[‘X_pca’][:, 0:ncomps] and ncomps is the number of components that will be used. |
true_label |
(list) ‘ground truth’ annotations or placeholder |
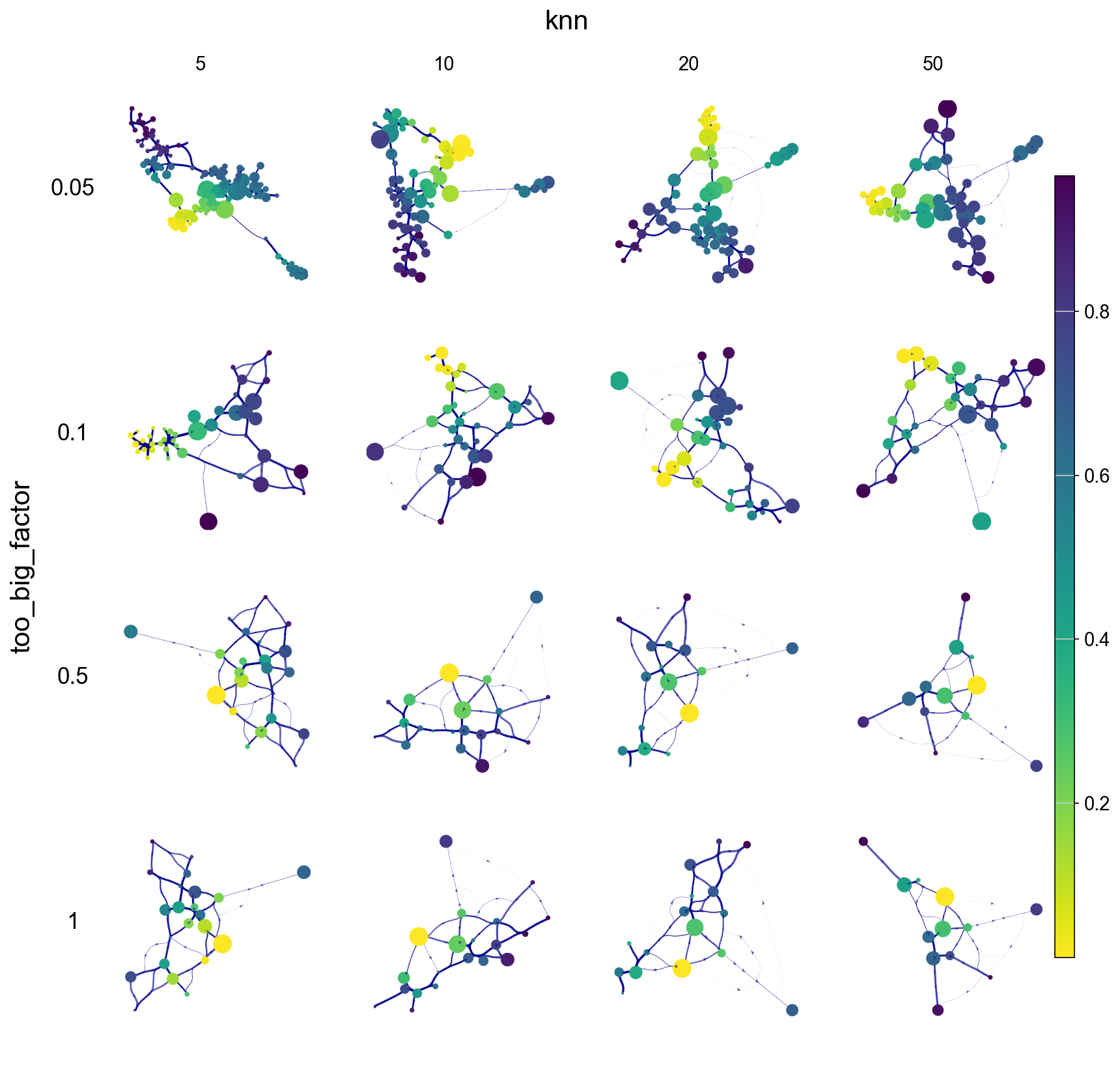
knn |
(optional, default = 30) number of K-Nearest Neighbors for HNSWlib KNN graph |
root_user |
(default is None) can be a list of strings, a list of int or None. When the root_user is set as None and an RNA velocity matrix is available, a root will be automatically computed. If the root_user is None and not velocity matrix is provided, then an arbitrary root is selected. If the root_user is [‘name_of_celltype_belonging_to_earlystage’] where the str corresponds to an item in true_label, then a suitable starting point will be selected corresponding to this group. If the root_user is in the form [678], where 678 is the index of the cell chosen as a start cell, then this will be the designated starting cell. It is possible to give a list of root indices and groups. [120, 699] or [‘traj1_earlystage’, ‘traj2_earlystage’] when there are more than one trajectories |
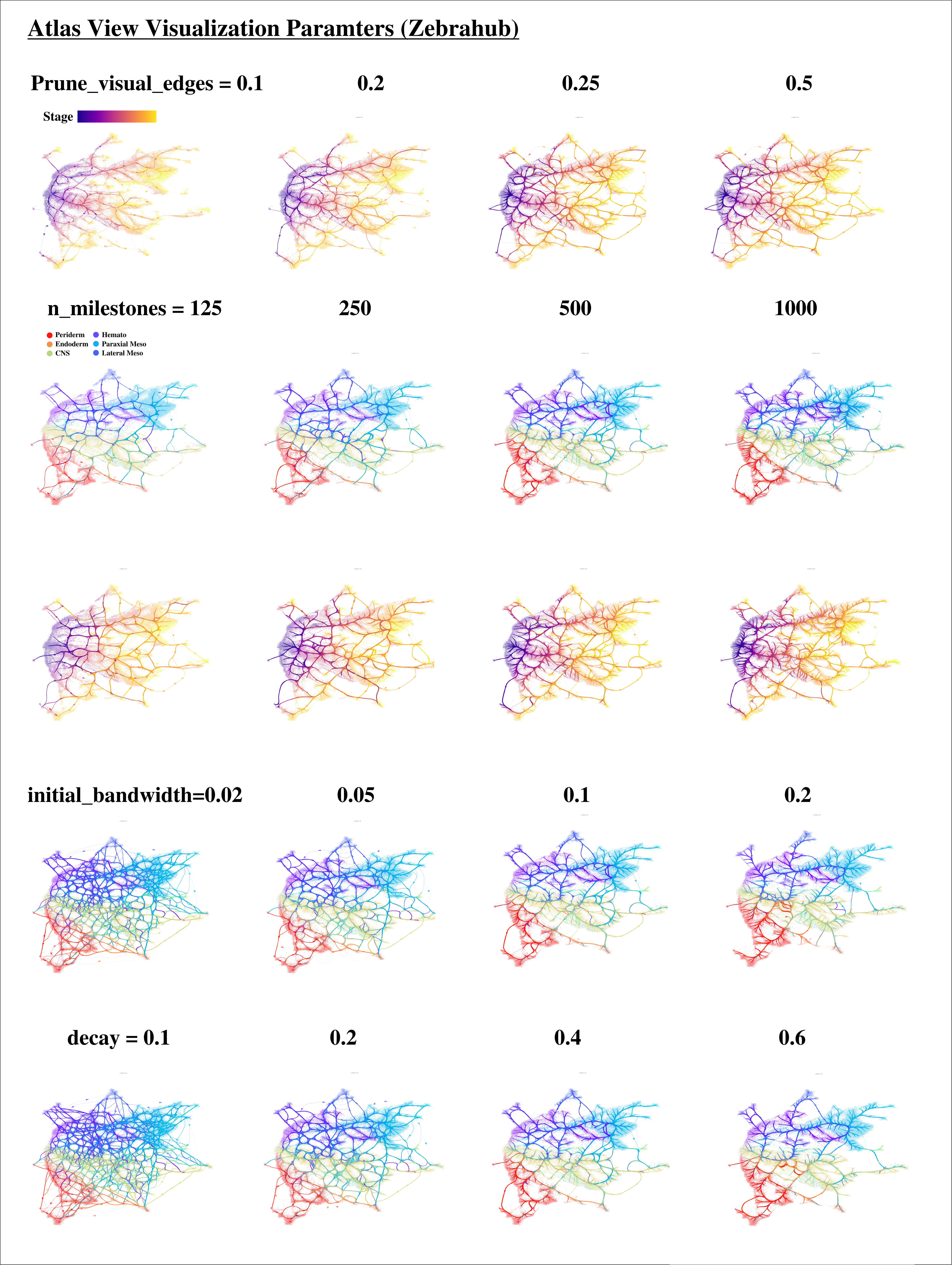
edgepruning_clustering_resolution |
(optional, default = 0.15) global level graph pruning for PARC clustering stage. Key tuning parameter. This threshold can also be set as the number of standard deviations below the network’s mean-jaccard-weighted edges. 0.1-1 provide reasonable pruning. higher value means less pruning. e.g. a value of 0.15 means all edges that are above mean(edgeweight)-0.15*std(edge-weights) are retained. We find both 0.15 and ‘median’ to yield good results resulting in pruning away ~ 50-60% edges |
edgepruning_clustering_resolution_local |
(optional, default = 1) Rarely needs to be tuned. local pruning threshold for PARC clustering stage: the number of standard deviations above the mean minkowski distance between neighbors of a given node. the higher the parameter, the more edges are retained |
resolution_parameter |
(optional, default = 1) Uses ModuliartyVP and RBConfigurationVertexPartition |
preserve_disconnected_after_pruning |
(optional, default = False) Cluster-graph pruning can occasionally cause fragmentation that can be repaired. However, if disconnected trajectories are believed to exist, then set this to True. |
cluster_graph_pruning |
(optional, default =0.15) Often set to the same value as the PARC clustering level of jac_std_global. To retain more connectivity in the clustergraph underlying the trajectory computations, increase the value |
memory |
(default = 5, reasonable ranges are 2-50) higher value means more memory, more retrospective/inwards randomwalk. memory = 1 runs the non-memory Via 1.0 mode. |
user_defined_terminal_cell |
(optional, default list = []) list of cell indices corresponding to terminal fate cells. |
user_defined_terminal_group |
(optional, default = list = []) list of group level labels corresponding to labels found in true_label, that represent cell fates. |
edgebundle_pruning |
(optional, default = None) This is automatically set to be the same as cluster_graph_pruning_std |
edgebundle_pruning_twice |
(optional, default = False) If the visualized cluster graph edges seem too busy, they can be further condensed by a second iteration of edge bundling by setting this to True. |
gene_matrix |
(optional matrix, not a numpy array, default = None) Only required when using RNA velocity to guide direction. Gene matrix not numpy array: adata.X.todense() |
velocity_matrix |
(optional matrix, default = None). Only required when using RNA velocity to guide direction. Matrix from scVelo with RNA velocities from: adata.layers[‘velocity’] |
velo_weight |
(optional, default = 0.5) #float between 0,1. the weight assigned to directionality and connectivity derived from scRNA-velocity |
too_big_factor |
(optional, default = 0.4) if a cluster exceeds this share of the entire cell population, then PARC will be run on the large cluster to increase granularity. |
x_lazy |
(optional, default = 0.95) 1-x = probability of staying in same node (lazy). Values between 0.9-0.99 are reasonable |
alpha_teleport |
(optional, default = 0.99) 1-alpha is probability of jumping. Values between 0.95-0.99 are reasonable unless prior knowledge of teleportation |
distance |
(optional, default = ‘l2’ euclidean) ‘ip’,’cosine’ |
random_seed |
(optional, default = 42) The random seed to pass to Leiden |
pseudotime_threshold_TS |
(optional, default = 30) Percentile threshold for potential node to qualify as Terminal State |
num_sim_branch_probability |
(optional), default = 500. Number of MCMCs run per terminal state. This can be safely reduced to 100 when computational resources are limited |
small_pop |
(optional, default = 10) Via attempts to merge Clusters with a population < 10 cells with larger clusters. |
Temporal Input Parameters
Input parameter |
Description |
|---|---|
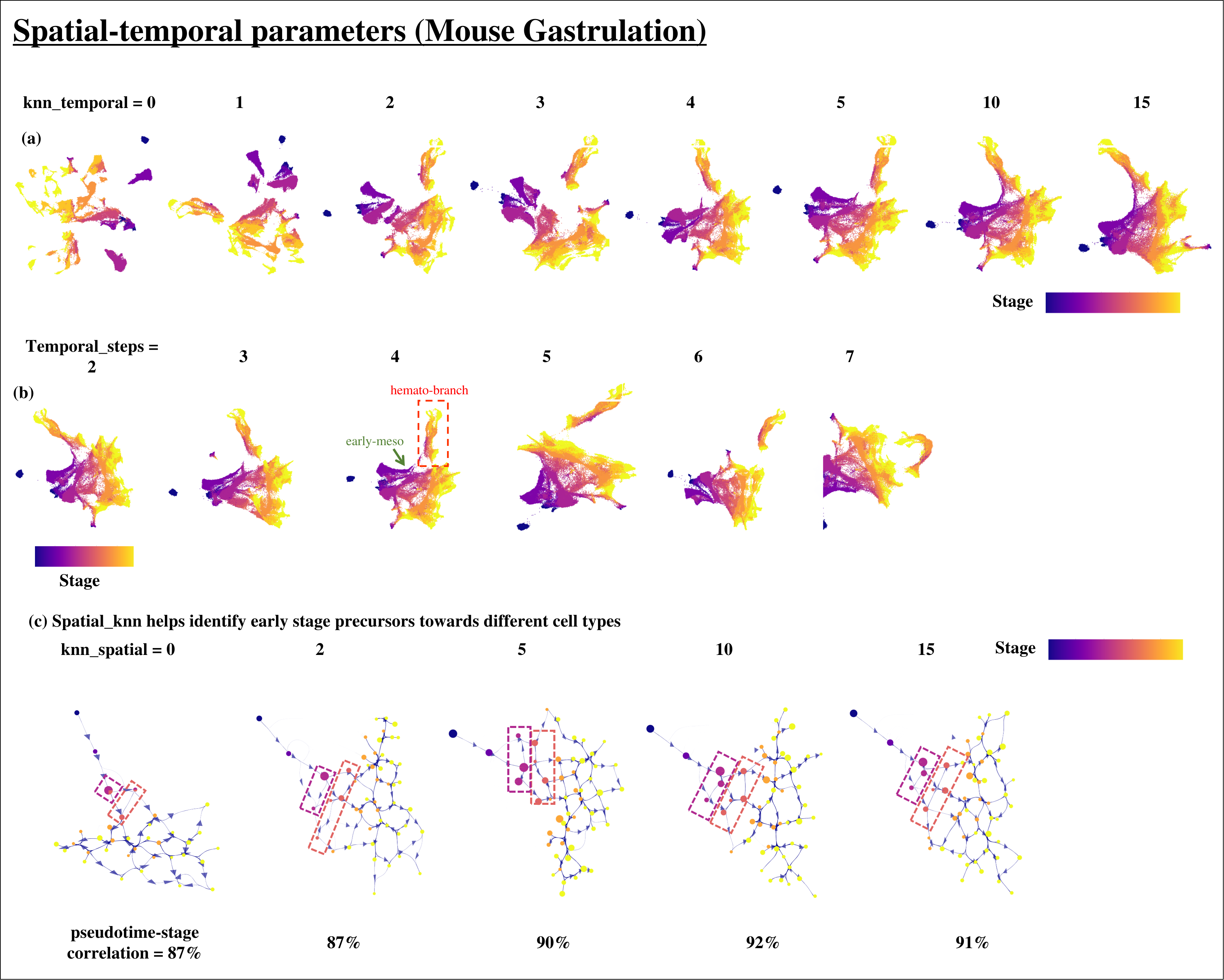
t_diff_step |
(optional, default = 1) Number of permitted temporal intervals between connected nodes. If time data is labeled as [0,25,50,75,100,..] then t_diff_step=1 corresponds to ‘25’ and only edges within t_diff_steps are retained |
time_series |
(optional, default False) if the data has time-series labels then set to True |
time_series_labels |
(optional, default None) list of integer values of temporal annoataions corresponding to e.g. hours (post fert), days, or sequential ordering |
knn_sequential |
(optional, default = 10) Number of knn in the adjacent time-point for time-series data (t_i and t_i+1) |
knn_sequential_reverse |
(optional, default = 0) Number of knn enforced from current to previous time point |
Spatial Input Parameters
do_spatial_knn |
(optional, default = False) Whether or not to do spatial mode of StaVia for graph augmentation |
|---|---|
do_spatial_layout |
(optional, default = 0.9) whether to use spatial coords for layout of the clustergraph |
spatial_coords |
(optional, default = False) np.ndarray of size n_cells x 2 (denoting x,y coordinates) of each spot/cell |
spatial_knn |
(optional, default = 15) number of knn’s added based on spatial proximity indiciated by spatial_coords |
spatial_aux |
(optional, default = []) a list of slice IDs so that only cells/spots on the same slice are considered when building the spatial_knn graph |
Attributes
Attributes |
Description |
|---|---|
labels |
(list) length n_samples of corresponding cluster labels |
single_cell_pt_markov |
(list) computed pseudotime |
single_cell_bp |
(array) computed single cell branch probabilities (lineage likelihoods). n_cells x n_terminal states. The columns each correspond to a terminal state, in the same order presented in the’terminal_clusters’ attribute |
terminal cluster |
(list) terminal clusters found by VIA |
super_cluster_labels |
Set this to v0.labels (clustering output of first pass “v0”) |
super_terminal_cells |
super_terminal_cells = via.get_loc_terminal_states(v0, data) |
full_neighbor_array |
full_neighbor_array = v0.full_neighbor_array. KNN graph from first pass of via - neighbor array |
full_distance_array |
full_distance_array = v0.full_distance_array. KNN graph from first pass of via - edge weights |
ig_full_graph |
ig_full_graph = v0.ig_full_graph igraph of the KNN graph from first pass of via |
csr_array_locally_pruned |
csr_array_locally_pruned = v0.csr_array_locally_pruned. CSR matrix of the locally pruned KNN graph |
Parameter Effects on VIA cluster-level trajectory graph
knn & too_big_factor effects colored by cell type and pseudotime

edgepruning_clustering_resolution & cluster_graph_pruning effects


Impact of Temporal and Spatial parameters on the StaVia Graph and Embeddings
Visualization parameters for the Atlas View